C·Prep Tissue RNA Extraction & Isolation Kit
The C·Prep Tissue RNA Extraction & Isolation Kit for animal tissues is a ready-to-use kit which comes with buffers formulated for use with animal tissues such as kidney, liver, lung, spleen, pancreas, etc. The protocol can be scaled to different input sizes but the standard input size is 8 mg of animal tissue.
Available Options:
| Purpose | RNA extraction, isolation, & purification from tissues such as liver, kidney, ect. |
|---|---|
| Format and Technology | Magnetic Beads; Carbon Technology |
| Processing Time | 15 min |
| Type of RNA Purified | Total RNA (includes ribosomal RNA, mRNA, and small RNAs) |
| Application | Northern, dot, and slot blotting, end-point RT-PCR, quantitative, real-time RT-PCR, next-generation sequencing |
| Processing | Manual or automated. Manual protocols use a water bath to release RNA, while automated protocols use robotic agitation to release RNA. |
| Main Sample Type | Tissue samples such as liver, kidney, and lung. For difficult samples such as bacterial biofilms we recommend the Phenol/Trizol kit. All C·Prep kits can be reconfigured into other C·Prep kits with changes to the buffers see the kit contents document under protocols for details. |
| Elution Volume | 30-100 µl |
| Sample Size | 8 mg of tissue is the standard input and reagent consumption scales with the amount of tissue. Less tissue may be used and the reagent use scales accordingly meaning that if less tissue is used the kit will be good for more than the listed number of reactions and vice versa. |
| Kit Storage | Room Temperature, one vial should be stored at 4° C |
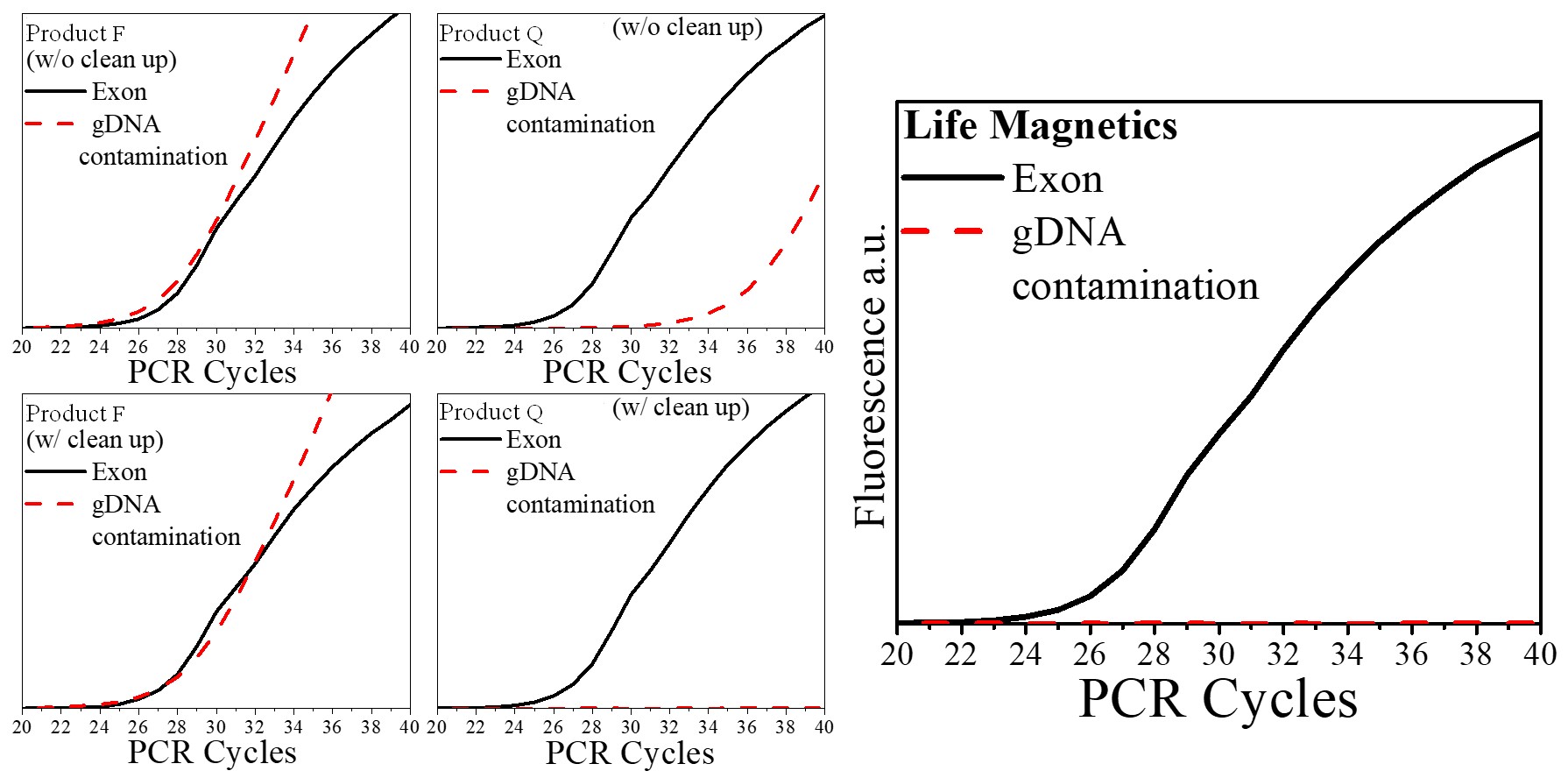
| Genomic DNA contamination | Carbon technology is >100x more selective than silica for RNA. In routine extractions gDNA contamination is typically ~0.5% – 1% of the sample making DNase cleanup unnecessary for routine PCR applications. If your application requires even higher purity, optimization of the protocol for your specific sample can result in <0.1% gDNA contamination. |
A comparison of extraction technologies using mouse kidney tissue is presented below. Technologies were compared by a third party at Wayne State University using the method presented in the following paper: “Padhi, B. K., Singh, M., Huang, N., & Pelletier, G. (2016). A PCR-based approach to assess genomic DNA contamination in RNA: Application to rat RNA samples. Analytical Biochemistry, 494, 49–51.” https://doi.org/10.1016/j.ab.2015.10.012
As compared to silica-based products, carbon has virtually no DNA contamination and does not require DNase cleanup. This is a huge advantage since DNase cleanup increases time and cost and damages RNA. During the COVID-19 pandemic, labs with at-home saliva tests cited lower DNA contamination in samples as an important factor for lower COVID-19 detection limits.
Li X, Frazier JA, Spahiu E, McPherson M, Miller RA. Muscle-dependent regulation of adipose tissue function in long-lived growth hormone-mutant mice. Aging (Albany NY). 2020;12(10):8766‐8789. doi:10.18632/aging.103380
Jones, L. B. et al. Effects of Pseudomonas aeruginosa on microglial-derived extracellular vesicle biogenesis and composition. Pathogens 8, 297 (2019). doi:10.3390/pathogens8040297
Shah, S., Brock, E. J., Jackson, R. M., Ji, K., Boerner, J. L., Sloane, B. F., & Mattingly, R. R. (2018). Downregulation of Rap1Gap: A Switch from DCIS to Invasive Breast Carcinoma via ERK/MAPK Activation. Neoplasia (New York, N.Y.), 20(9), 951–963. doi:10.1016/j.neo.2018.07.002
- “The main drawback from my current RNA isolation protocol is the numerous centrifugation steps and the relatively low volume of RNA yield in the end. Life Magnetics provides a higher yield of RNA from samples and eliminates the use of centrifugation steps.”
– Kendra Royston, Ph.D. at University of Chicago - “Life Magnetics has a new technology, which is worth trying and their biggest advantage is skipping DNase step.”
– Quan Taihao, Ph.D. and Qin Zhaoping at University of Michigan, Dermatology Department - “It is easy to use and gives high quality and a greater yield at a much-reduced price. Also, the stability of RNA with the beads at room temperature before the final extraction step is a big plus.”
– Sabita Saldanha, Ph.D. at Alabama State University - “This system is significantly easier than Trizol because it can be done on the bench top, rather than a fume hood. It is of comparable ease to Qiagen systems. A potential advantage that this kit has over a Qiagen system is that it can be scaled more easily.”
– Joe Endicott, Ph.D. at University of Michigan, School of Medicine Ann Arbor - “With Life Magnetics Carbon-Based RNA extraction kit, it was nice to switch things up and try a different protocol. The extraction method generated great RNA purity and yield.”
– Kayla Lewis, Ph.D. at the University of Alabama at Birmingham - “When I used the Life Magnetics IsoMag Carbon Based RNA purification kit, I was surprised how easy protocol was to follow and how much more RNA was extracted. The major advantage of this kit over other commercial kits is high RNA yield with no additional DNase cleanup steps.”
– Seema Shah, Ph.D. at Wayne State University
Safety data sheets for all materials included in kit: